
Apache Spark is the top framework for data analysis and processing. I often find that data scientists and data engineers get frustrated using Spark and PySpark due to perceived slowness, performance, etc.

This perception is wrong and the root cause of their issue is job configuration not using best practices or application design. Let's look into a couple of the common issues faced and how to fix them.
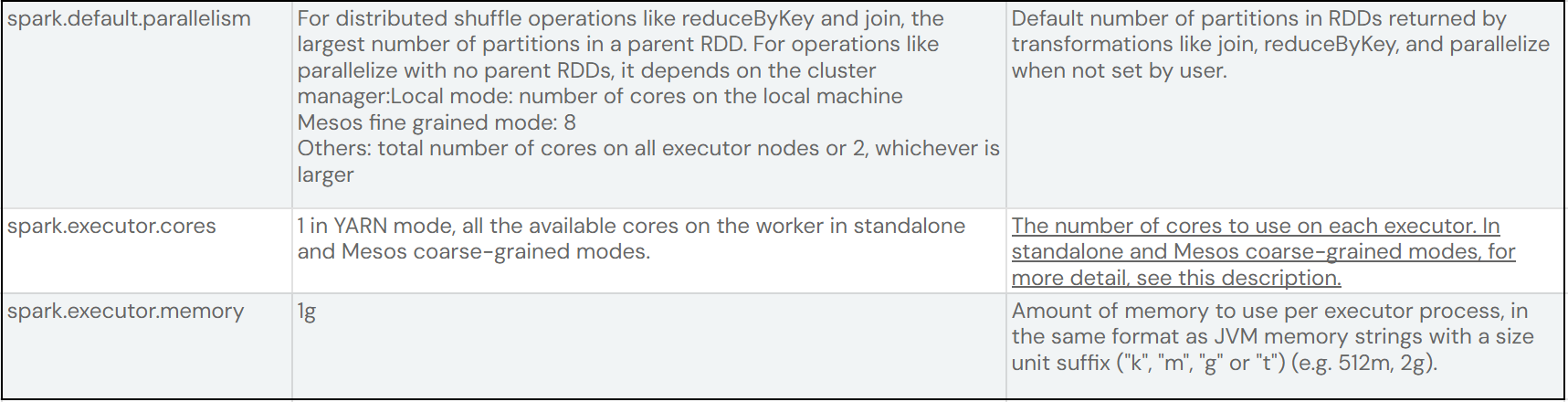
These are the two configuration properties we'll look at.
Executors are jvms that live on worker nodes in the cluster. The primary configuration items are the core and memory. Settings these incorrectly will cause performance issues with your job. In order to properly configure your executors you need to understand the infrastructure scaling capabilities (static, dynamic). If your cluster does autoscale then understand the maximum resources you are able to use.
We'll look at a couple scenarios for static and dynamic clusters.
You have a 20 node cluster, each node is 12 core, 48 GB ram.
When configuring the executors you want to align the configuration with the node. I've seen engineers set the executor cores to 16 and memory to 96 GB. This is equivalent to spinning up a jvm to use more resource than are available on the host. Also setting the executor cores to 5 does not make sense as 12 is not equally dividable by 5. Perhaps a configuration of 2 core, 8gb ram is proper. You can fit 6 executors (2 core * 6 executor = 12 cores and 8GB ram * 6 executors = 48GB ram).
If this is a managed cluster then sometimes the administrators will cap the cpu & ram, so you don't have the ability to override it. At that point you are just calculating total executors as your only configurable parameter.
Takeaways
Much like static infrastructure you need to align with the server capacity. You typically have the ability to spaw a larger number of executors but the servers are usually smaller due to cost. 1 core, 2gb ram executors are seen often when elastic servers are 4 core, 16gb ram. If you have an application that needs a significant amount of memory you might need to use 1 executor at 4 core and 16gb ram in order to process the data.
We will now look at the relationship between spark.default.parallelism and spark.executor.cores
The default value for spark.default.parallelism is 200. So if we have 20 executors and set the core count to 3 we get 60 (20 executors * 3 cores = 60) tasks. So we can process 60 items in parallel.
Let's say the dataset is large and will require 900 task to read it. Our job will process 60 tasks concurrently, when complete 60 more item and continue in waves of 60, but with 200 parallelism it will do 3 waves of 60 for 180 tasks but then the last set of tasks is 20, which means the other 40 tasks will watch the final 20 run to completion before they move on to the next 200 tasks.
So the executor cores needs to be dividable into the parallelism. We can set shuffle to 180 or 360. If we set it to 180 then we get 3 wave os 60 task, then 180 goes into 900 nicely 5 times to equal 900. So we get 3 waves of 60 then 5 equal passes so that we maximize resource and use every core throughout the job, no cores wasted = fast execution = no money wasted.
These are simple things we can do once, and the saving will last throughout the life of the job. Imagine the cost of not doing this analysis on a job that runs every 5 mins, 12 times and hour, 288 times a day and 105,120 times per year....add up the cost then multiply it by the total number of jobs your have (hundreds, thousands)?
You can see the benefits of doing the analysis.
Engineers often need to join datasets to accomplish a task. There is one critical mistake that engineers routinely make when joining datasets. It is best practice to repartition your datasets by the same join key in order to have optimal performance.
Assume you have two datasets, customers and orders. The join key is the customer id. We want to repartition both datasets by customer_id so that the same customer_id will be placed into one partition.
Assuming we have 200 partitions, if customer_id 1234 is spread across all the partitions, 5 in one part, 3 in another, 0 in yet another and 24 in another then all the partitions must be read into memory in order to perform the join.
If we do a customers_df.repartition('customer_id', 10) then records that have customer_id of 1234 will all be contained in the same partition. If we do the same for the order datasets (orders_df.repartition('customer_id', 10)) then each customer partition can be joined directly to each order partition eliminating a shuffle and significantly improving the join performance.
Be sure to understand the difference between actions and transformations. If you don't cache after an action then the next action will go back to the start of the DAG and read the data from disk again. I have seen engineers state that "spark ingestion is slow" when in fact they were reading the same dataset from disk 5 times because they chained together 5 actions without caching at any point in their program. If you only have one action then you don't need to cache, if you have more than one you should cache and repartition by join keys after the read from disk action is complete.
When writing data to any data source it is important to understand that each partition is written to target by an executor task. So in order to speed up the writing of data you need to increase the number of partitions in the dataframe and verify that you have enough executor and tasks available to write all the partitions concurrently.
When calling the map operation on a rdd (rdd.map) or a dataframe (dataframe.rdd.map) you need to be aware that the map operation executes on each partition. So make sure that only required operations take place in the map functions. I have seen engineers write a file to S3 within the map function which significantly slowed down the performance of the function. It would better to place the s3 write outside the map function, so write the resulting rdd once instead of chunks of the rdd during execution. You could also use mapPartitions and write the data for collections of RDDs vs one at a time.